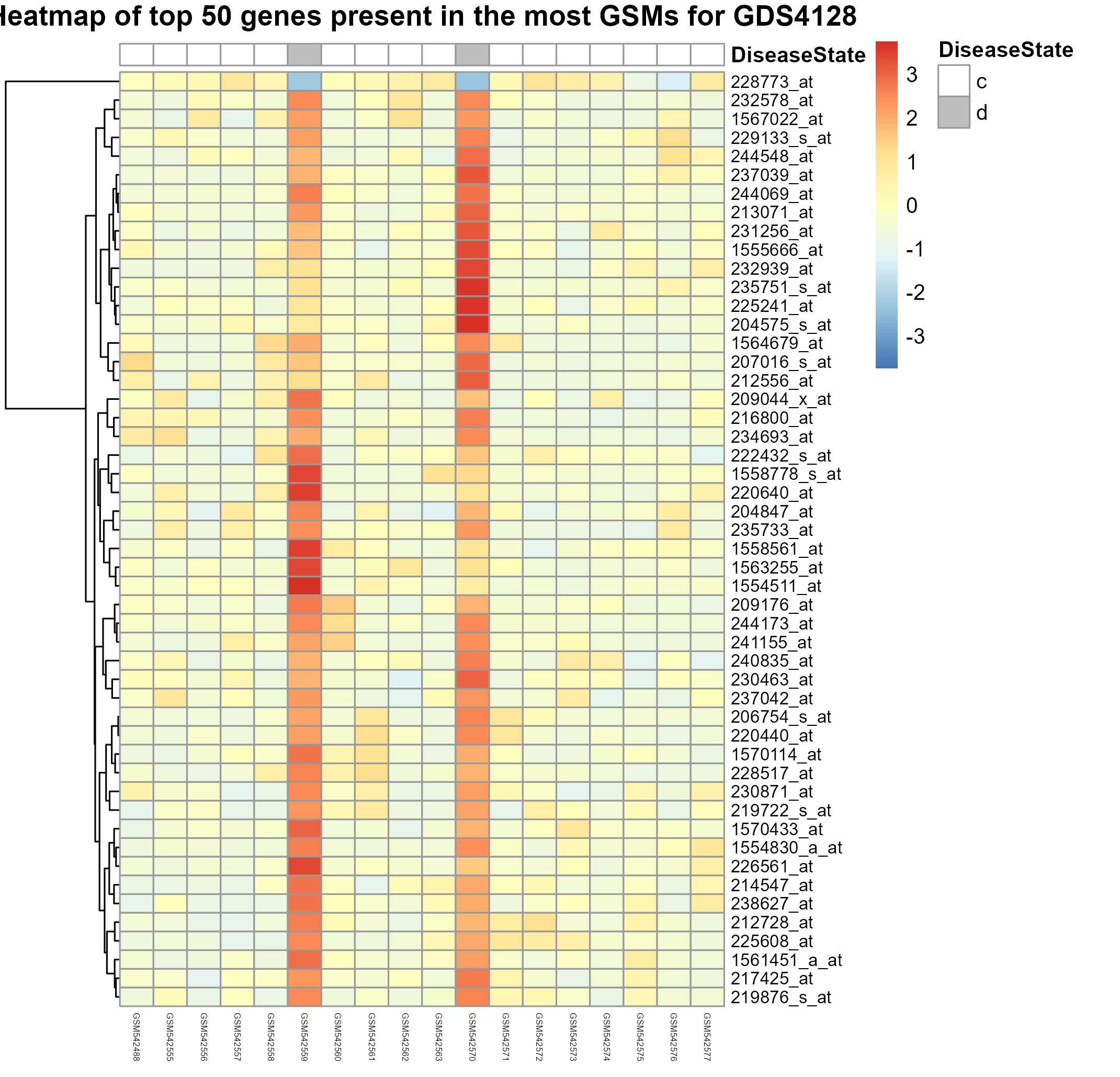
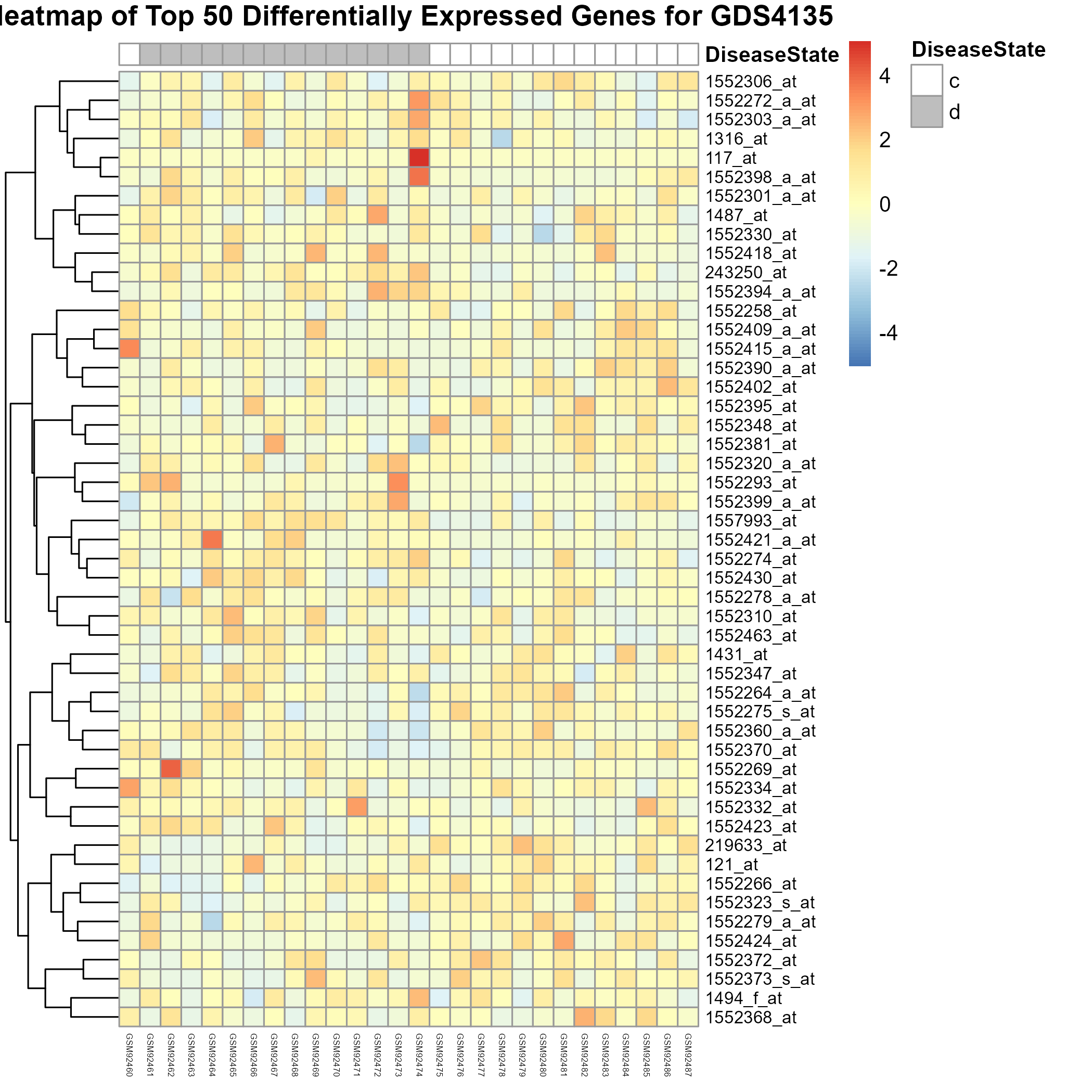
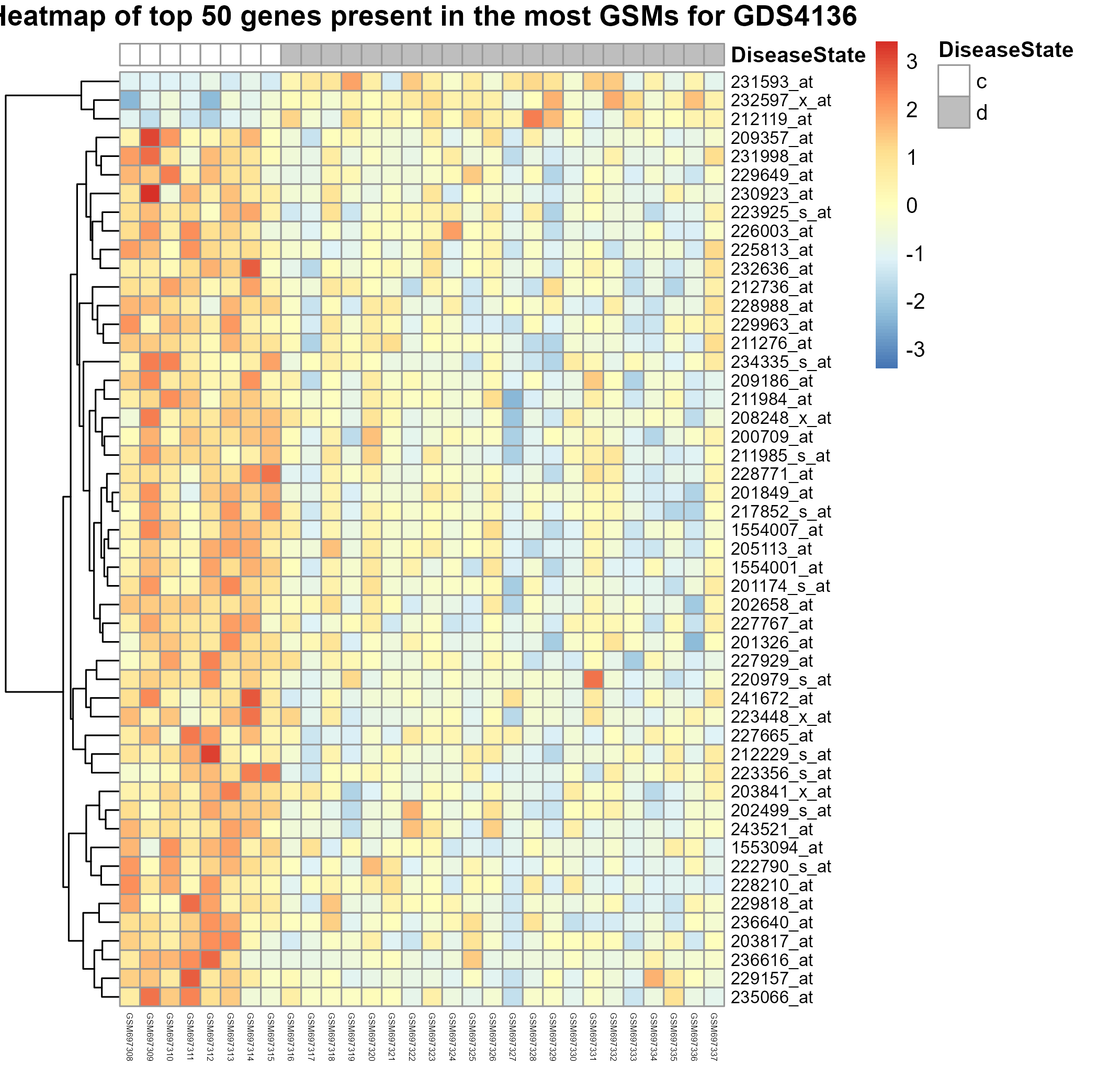
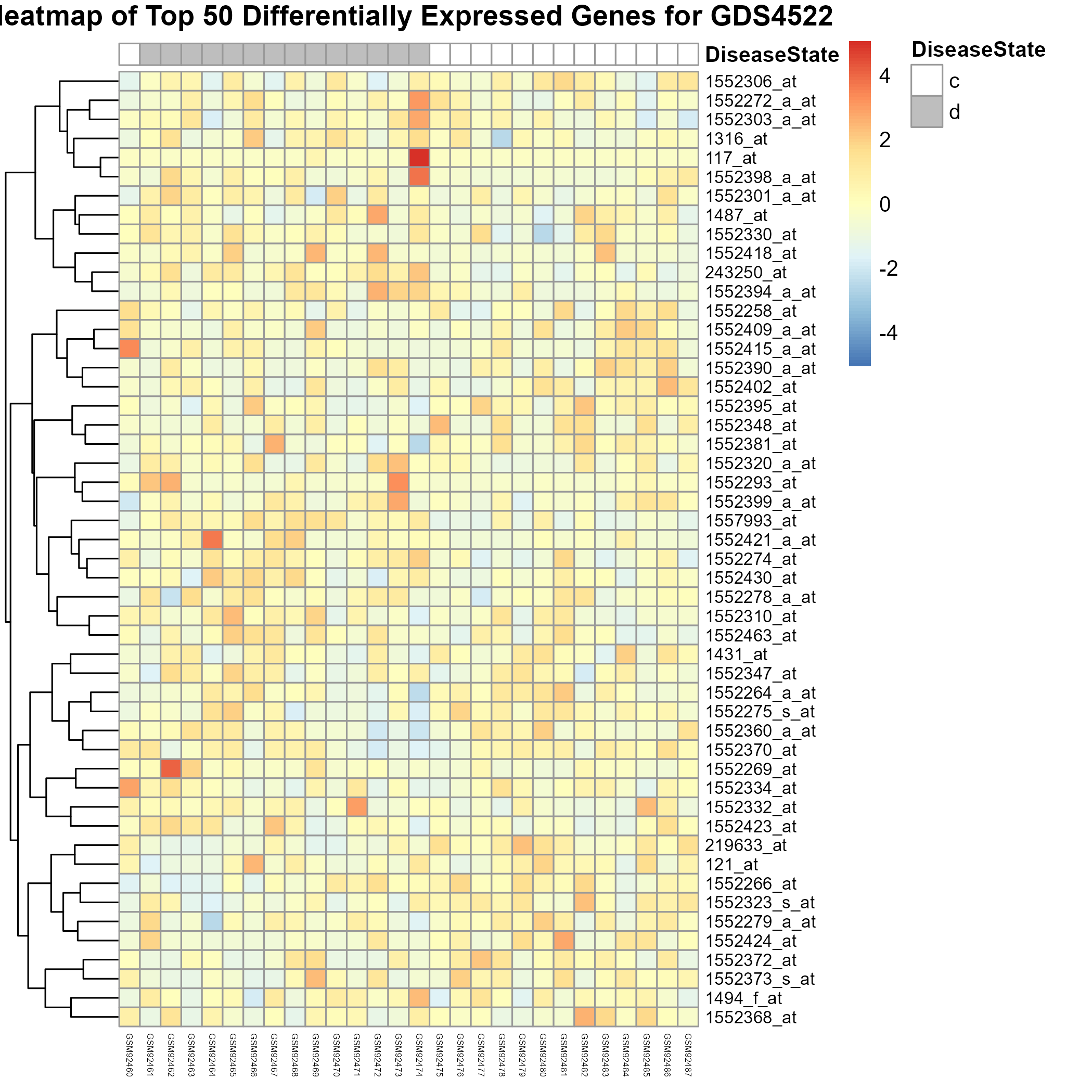
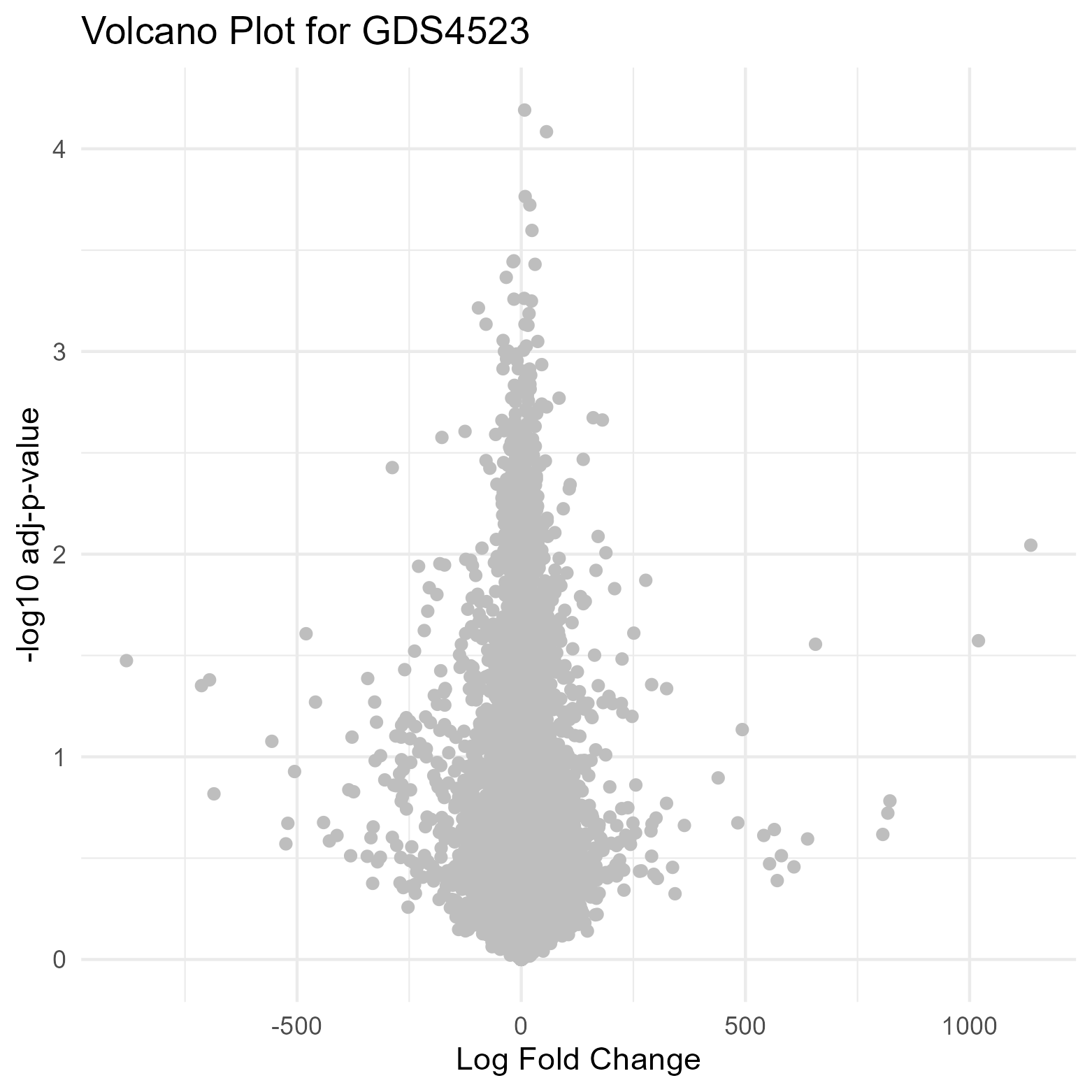
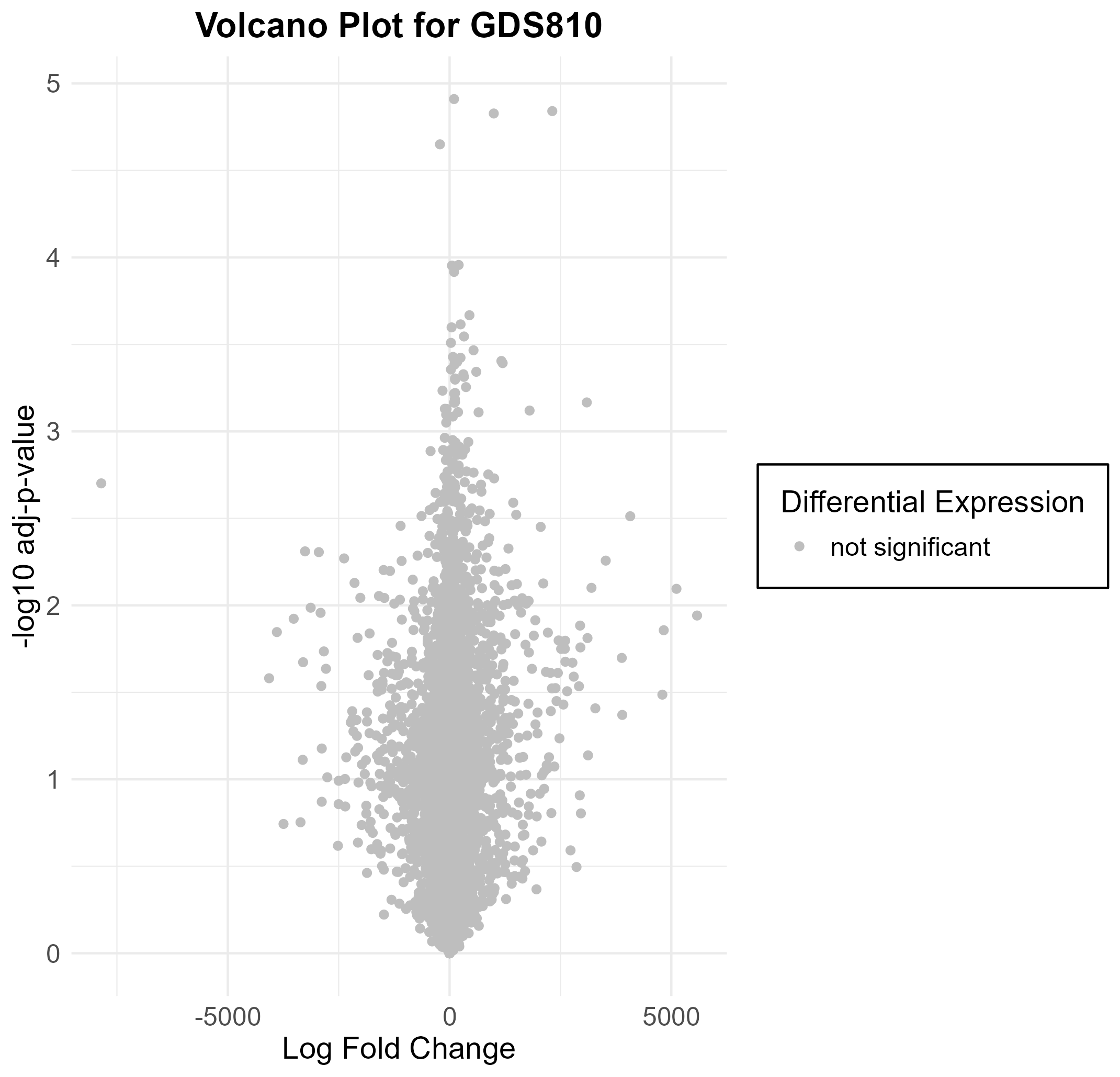
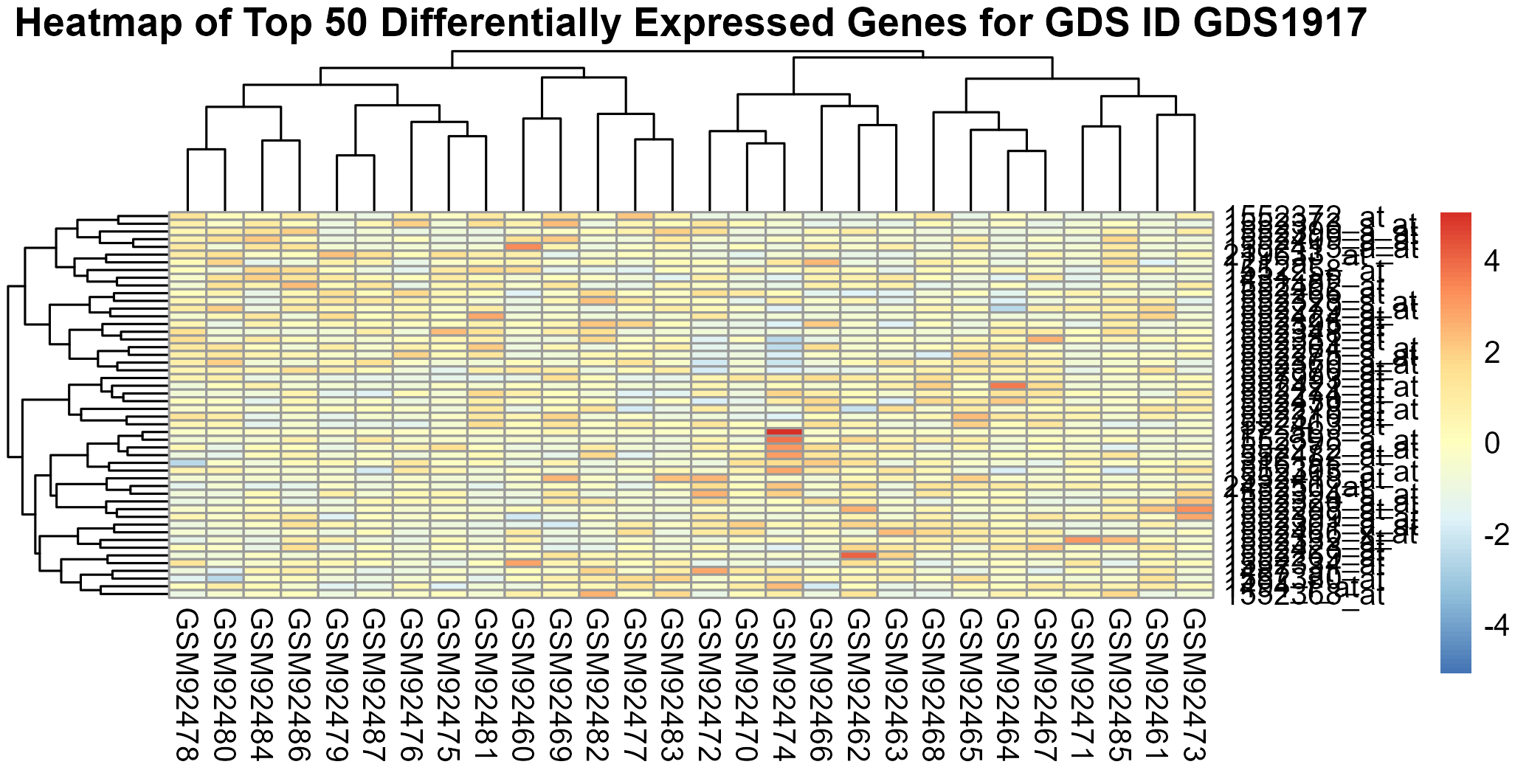
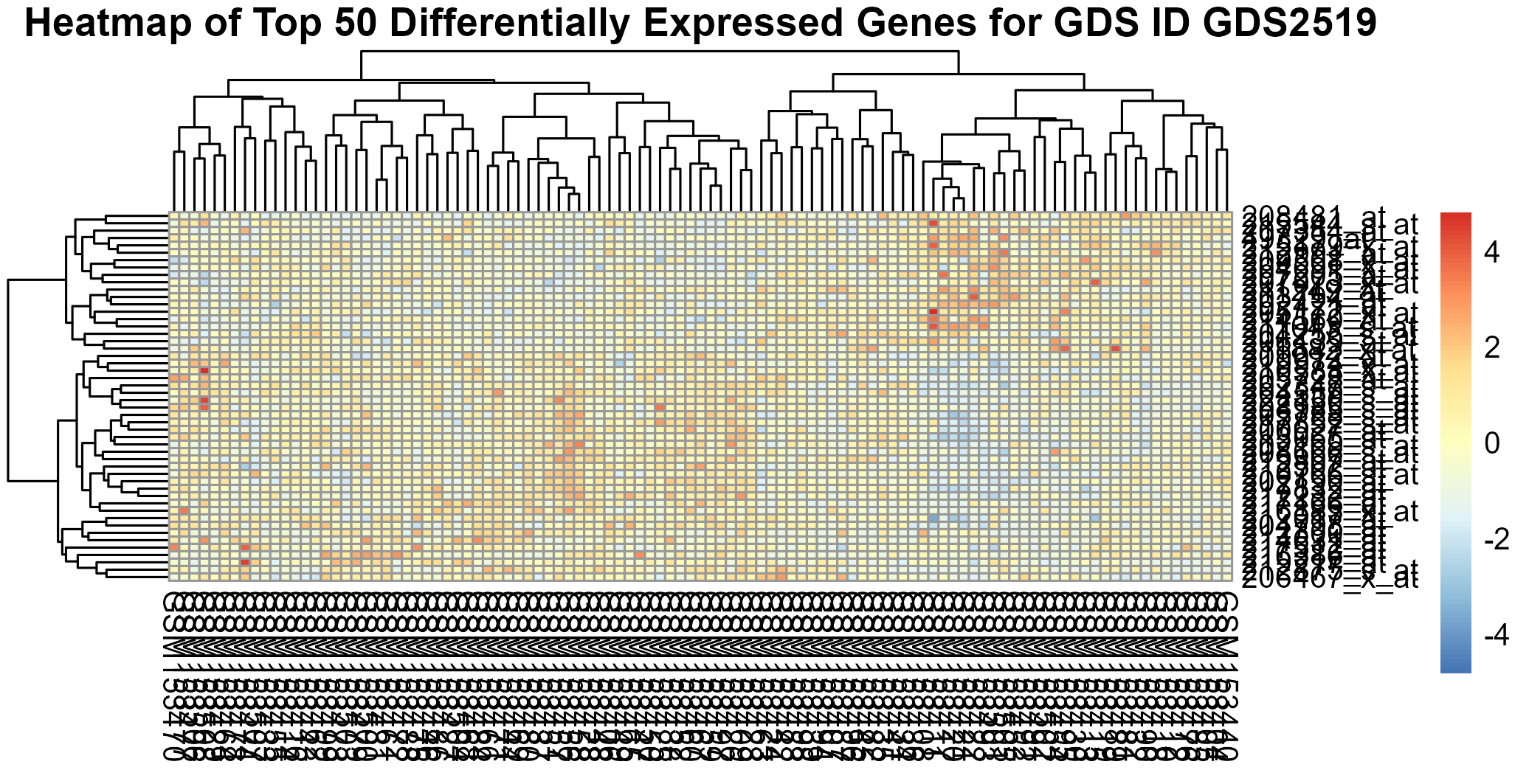
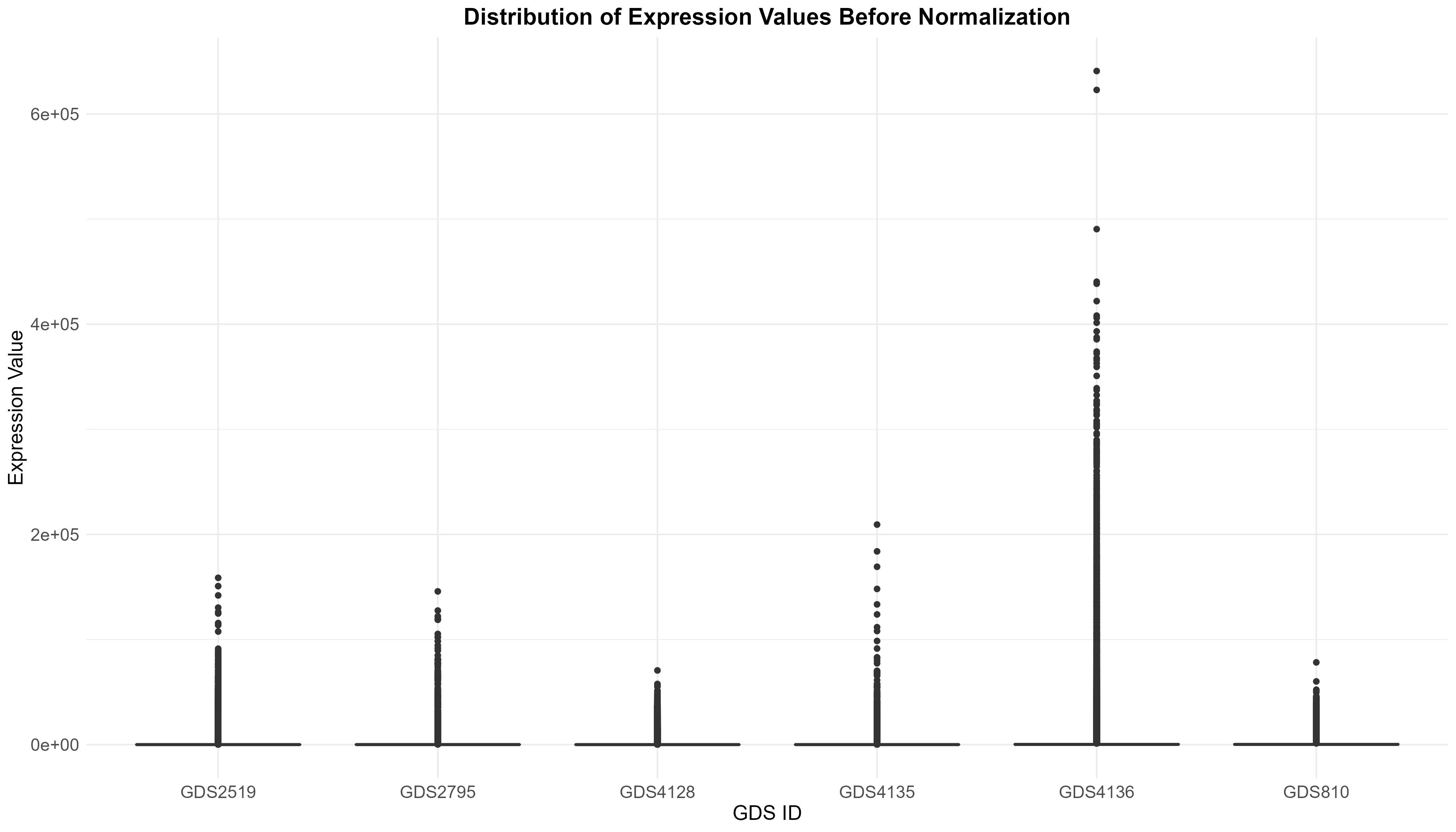
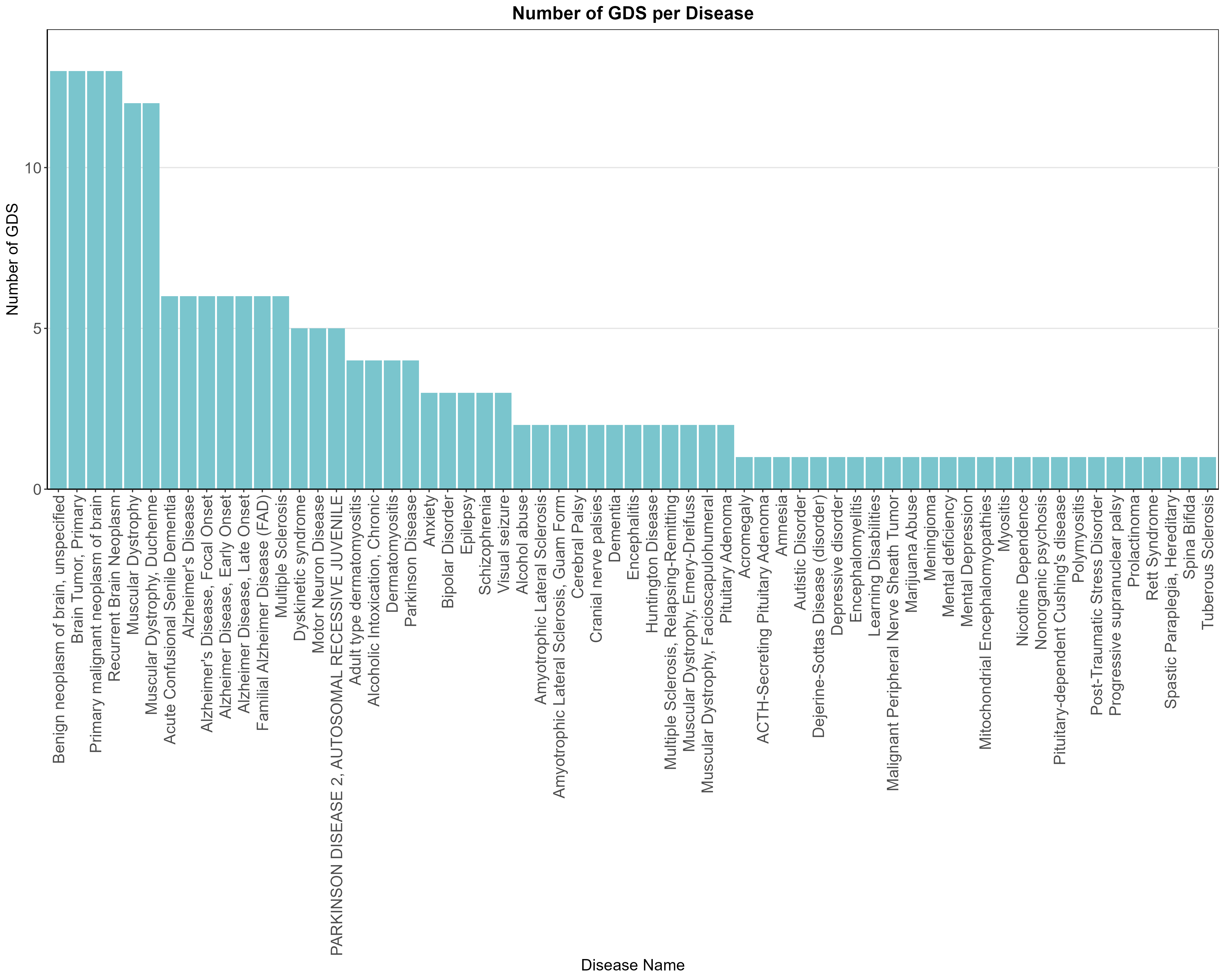
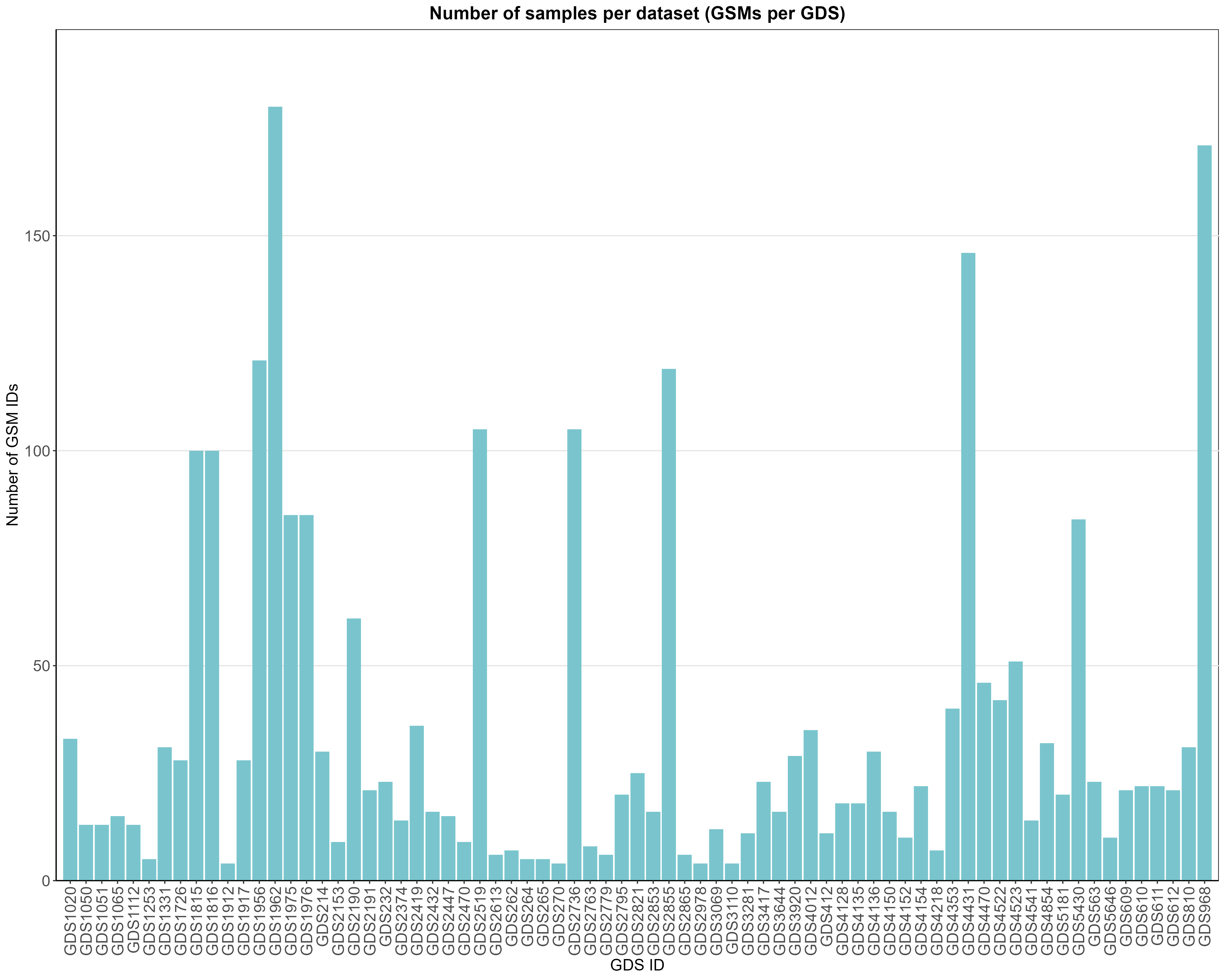
Uploading data_analysis correctly
Showing
This source diff could not be displayed because it is too large. You can view the blob instead.
90.4 KB
272 KB
269 KB
275 KB
287 KB
298 KB
314 KB
298 KB
302 KB
324 KB
182 KB
155 KB
158 KB
158 KB
160 KB
211 KB
121 KB
185 KB
This source diff could not be displayed because it is too large. You can view the blob instead.
247 KB
357 KB
213 KB
221 KB
238 KB
59.7 KB
111 KB
101 KB
101 KB
62.3 KB
114 KB
105 KB
105 KB
59.3 KB
110 KB
98 KB
103 KB
57.7 KB
95.3 KB
101 KB
95.5 KB
60.7 KB
115 KB
108 KB
110 KB
60.2 KB
123 KB
98.3 KB
98.8 KB
126 KB
136 KB
717 KB
185 KB